Brettanomyces bruxellensis: overview of the genetic and phenotypic diversity of an anthropized yeast.
Jules Harrouard, Chris Eberlein, Patricia Ballestra, Marguerite Dols-Lafargue, Isabelle Masneuf-Pomarede, Cécile Miot-Sertier, Joseph Schacherer, Warren Albertin.
Molecular Ecology. 2022. doi: 10.1111/mec.16439
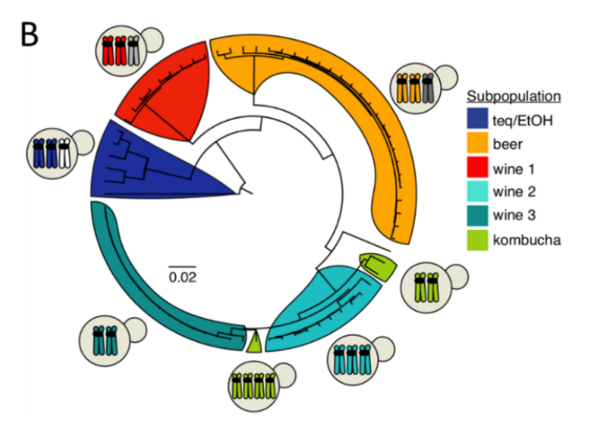
Human-associated microorganisms are ideal models to study the impact of environmental changes on species evolution and adaptation because of their small genome, short generation time, and their colonization of contrasting and ever-changing ecological niches. The yeast Brettanomyces bruxellensis is a good example of organism facing anthropogenic-driven selective pressures. It is associated with fermentation processes in which it can be considered either as a spoiler (e.g. winemaking, bioethanol production) or as a beneficial microorganism (e.g. production of specific beers, kombucha). Besides its industrial interests, noteworthy parallels and dichotomies with Saccharomyces cerevisiae propelled B. bruxellensis as a valuable complementary yeast model. In this review, we emphasize that the broad genetic and phenotypic diversity of this species is only beginning to be uncovered. Population genomic studies have revealed the co-existence of auto- and allotriploidization events with different evolutionary outcomes. The different diploid, autotriploid and allotriploid subpopulations are associated with specific fermented processes, suggesting independent adaptation events to anthropized environments. Phenotypically, B. bruxellensis is renowned for its ability to metabolize a wide variety of carbon and nitrogen sources, which may explain its ability to colonize already fermented environments showing lownutrient contents. Several traits of interest could be related to adaptation to human activities (e.g. nitrate metabolization in bioethanol production, resistance to sulphite treatments in winemaking). However, phenotypic traits are insufficiently studied in view of the great genomic diversity of the species. Future work will have to take into account strains of varied substrates, geographical origins as well as displaying different ploidy levels to improve our understanding of an anthropized yeast’s phenotypic landscape.